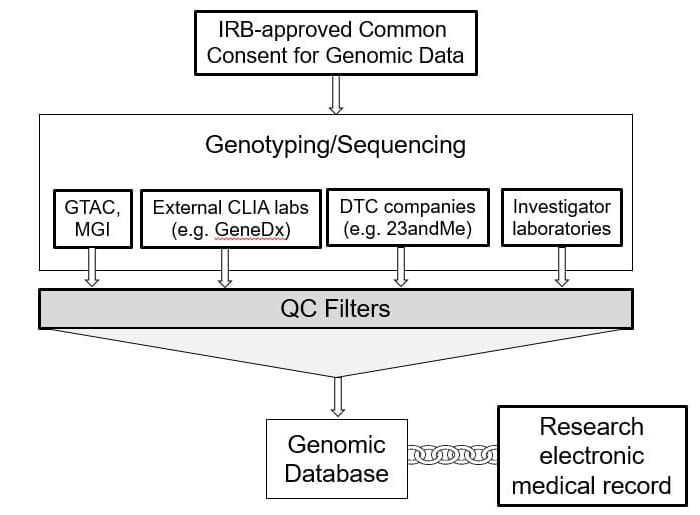
What is the “one protocol one consent”?
The “one protocol one consent” is an IRB approved protocol and consent that will permit the Institute of Informatics (I²) to link participant or patient genetic data that you collect to the research copy of their BJC electronic health record.
What is the purpose of the “one protocol one consent”?
The goal of this standardized protocol and consent is to reduce the burden for individual researchers wishing to make progress in genomics and precision medicine, both for their teams and the wider research community. The standardized consent incorporates best-practices language for informing participants about genomics research and potential return of research results. It has been approved by the IRB for use by all WashU/BJH researchers and clinicians. Use of this protocol and consent as a companion to the consent for your own study or in your clinical care will facilitate future analyses for your team and lead to a large campus-wide resource for long-term research in precision medicine.
How do I get started?
Researchers and clinicians interested in using the “one protocol one consent” should contact the Precision Health Navigator Tricia Salyer at salyerp@wustl.edu to be added to the study’s protocol and provided instructions for use.

ICTS UK Biobank Genomic Repository
To facilitate genomic research, the ICTS Precision Health Function has established the ICTS UK Biobank Genomic Repository. This Repository includes genomic data for 500,000 participants from the UK Biobank and has been enriched through annotation by the McDonnell Genome Institute (MGI). Using the ICTS UK Biobank Genomic Repository allows researchers to access a curated and enriched version of the data. The Repository is stored with Research Information Services (RIS) and is available for access by all approved UK Biobank users at WUSM.
The vast majority of UK Biobank data is accessible only within their cloud environment. Access to the UK Biobank data is available for a fee and cloud resources are charged when used.
UK Biobank offers free credits to early career researchers (within 4 years of degree, 4 years of starting their first academic appointment or student status). More than $40,000 available per user. This program is available whether or not your PI has access to UK Biobank. Enrollment gives you access to full genomes, exomes, imaging, proteomes and more.
If you have an approved UK Biobank project and would like access to the ICTS UK Biobank Genomic Repository for yourself and your research team, please email the Precision Health Administrator Debra Warren at debrawarren@wustl.edu.
Details about the ICTS UKB Repository can be found in the documents below.
- What is in the ICTS UKB Repository (pdf)
- How to Access the Repository (pdf)
- Access all UK Biobank datasets through the DNAnexus research analysis platform page
- Use the community page to access content for learning and troubleshooting the analysis platform
- Who at Washington University is publishing with this dataset? View the list here (updated monthly, last update July 2024) To request list edits contact ictsprecisionhealth@wustl.edu
The database of Genotypes and Phenotypes (dbGaP) was developed to archive and distribute the data and results from studies that have investigated the interaction of genotype and phenotype in humans. Investigators can request access to dbGaP datasets for approved research projects, or they can deposit their own data.
The documents below provide detailed guidance for researchers who wish to request and submit data to dbGaP.
Contact Jenny McKenzie at j.mckenzie@wustl.edu with any questions about these documents.
The Greater Plains Collaborative (GPC) is a network of 13 leading medical centers in 8 states committed to a shared vision of improving healthcare delivery through ongoing learning, adoption of evidence – based practices, and active research dissemination. The network brings together a diverse population of over 34 million patients. More information can be found on their website: https://gpcnetwork.org/ Researchers can submit data requests for patient counts, de-identified data or limited data at the GPC Query and Data Request form.
The All of Us Research Program, part of the National Institutes of Health, is building one of the largest biomedical data resources of its kind. The All of Us Research Hub stores health data from a diverse group of participants from across the United States.
There are three tiers of data available. The public tier contains aggregate data with identifiers removed. Registered tier data contains electronic health records (EHRs), wearables, surveys, and physical measurement data. Controlled tier data contains genomic data in the form of whole genome sequencing (WGS) and genotyping arrays, including previously suppressed demographic data fields from EHRs and surveys.
Approved researchers can access All of Us data and tools to conduct studies to help improve our understanding of human health.
Who at Washington University is publishing with this dataset? View the list here (updated monthly, last update July 2024) To request list edits contact ictsprecisionhealth@wustl.edu
To learn more about the All of Us Research Program and the resources available to researchers, please review the following resources:
- The Research All of Us Website
- Video on conducting research with the All of Us Researcher Workbench
- Researcher Testimonials
- All of Us Research Program
- All of Us Researcher Workbench
- How to Register
- Find out if your Institution has a DURA
- Attend Office Hours with All of Us
Want to learn more about All of Us?
- See where the program is on the Journey to one million participants
- See the work of current active users in the project directory
- See how All of Us is making an impact through publications
- Check the Data Browser to see what data types and aggregate counts are currently available
- Subscribe to the Research Hub Newsletter
- Find funding opportunities
- Locate and read additional documentation in the User Support Hub
- Learn more about computational costs
Digital Commons Data@Becker is a repository for faculty, staff, students and trainees at Washington University School of Medicine to share their data and supporting files in compliance with funder and publisher policies.
To start the data sharing process, submit the Data Management and Sharing Consultation Request form.
For more information about our services in this area please visit Becker Medical Library’s Data Management and Sharing site or contact Seonyoung Kim and Xing Jian at BeckerDMS@wustl.edu.
What is WashU Research Data (WURD)?
WURD is a formal research data repository that supports research data sharing by providing:
- DataCite metadata
- Digital Object Identifiers (DOIs)
- Open Researcher and Contributor ID (ORCiD)
- Research Organization Registry (ROR)
- Award Information
- Integrated curatorial review for data quality
- Github integration
- Long-term preservation
- FAIRness review
- Indexed for search engines, integrated with discovery tools
Who can use WURD?
All Washington University faculty, students, staff, and other authorized University affiliates can use WURD and related Libraries’ services for data curation and sharing. Washington University Libraries provides data curation and sharing services to support Washington University researchers and scholars across all WashU schools and campuses.
Choosing a Data Repository
WURD is an institutional repository, and you may also consider domain and generalist repositories available to you. Becker Medical Library also offers repository services tailored for School of Medicine affiliates. More information on their services can be found on the Becker Medical Library Data Management and Sharing website. Library staff on either campus are available to assist you in choosing the most appropriate repository option.
How to Get Started
You can start your data deposit today at the WURD website.
Questions?
Contact researchdata@wustl.edu.